library(tidyverse) # for dplyr, ggplot2 packagesThis is the third of several posts related to McCarrick et al. (2022).
Introduction
McCarrick et al. (2022) examined the population dynamics of Yellowstone Cutthroat Trout (Oncorhynchus clarkii bouvieri) in Henrys Lake, Idaho over a nearly two decade period. Their Figure 4 showed mean (with confidence interval) relative weight (Wr) for various length categories of Cutthroat Trout across years. I use ggplot2 to recreate that figure here.
The following packages are loaded for use below. A few functions from each of lubridate, FSA, plyr, scales, gghrx, lemon, and ggtext are used with :: such that the entire packages are not attached here.
McCarrick et al. (2022) computed what I am calling an overall PSD1 and what are called “incremental” PSD indices. I assume that these are familar to most fisheries scientists, but they are described in more detail in Chapter 8 of Ogle (2016).2
1 This is the most common PSD measure.
2 Also see Blackwell et al. (2000).
Data Wrangling
Individual Fish Data Frame
McCarrick et al. (2022) provided raw data for Figure 2 as an Excel file in their Data Supplement S1. The same data wrangling, up to where catch-per-unit-effort was calculated, used in this previous post is used here3 and, thus, will not be discussed in detail.
3 And in this post
dat <- readxl::read_xlsx("../2023-3-22_McCarricketal2022_Fig2/download.xlsx",
na=c("","??","QTY =","QTY=","UNK","NO TAG"),
col_types=c("date","numeric","text",
"numeric","numeric","text")) |>
mutate(year=lubridate::year(Date),
year=ifelse(year==1905,2002,year)) |>
filter(!is.na(year)) |>
select(species=Species,year,length,weight) |>
mutate(species=case_when(
species %in% c("YCT","Yct") ~ "YCT",
species %in% c("UTC","CHB","CHUB") ~ "UTC",
TRUE ~ species
)) |>
filter(species %in% c("YCT","UTC")) |>
mutate(species2=plyr::mapvalues(species,
from=c("YCT","UTC"),
to=c("Cutthroat Trout","Utah Chub"))) |>
mutate(gcat=FSA::psdAdd(len=length,species=species2))
FSA::headtail(dat)#R| species year length weight species2 gcat
#R| 1 YCT 2002 150 NA Cutthroat Trout substock
#R| 2 YCT 2002 160 NA Cutthroat Trout substock
#R| 3 YCT 2002 160 NA Cutthroat Trout substock
#R| 19900 YCT 2020 391 550 Cutthroat Trout quality
#R| 19901 YCT 2020 284 229 Cutthroat Trout stock
#R| 19902 YCT 2020 440 853 Cutthroat Trout qualityThe relative weigt for each fish needs to be added to this data frame, preferably with wrAdd() from FSA. Note, however, that there are two standard weight equations for Cutthroat Trout, one for lentic and one for lotic populations. We want to make sure to use the one for lentic populations here and, thus, cannot use the species name found in the data frame within wrAdd(). Note below that the data frame was also reduced to just Cutthroat Trout for which a weight was recorded, and the two species variables were dropped (just to simplify the output).
dat <- dat |>
filter(species=="YCT",!is.na(weight)) |>
mutate(wr=FSA::wrAdd(weight,length,spec="Cutthroat Trout (lentic)")) |>
select(-starts_with("species"))
FSA::headtail(dat)#R| year length weight gcat wr
#R| 1 2004 174 54 substock 102.34315
#R| 2 2004 227 130 stock 108.45450
#R| 3 2004 305 380 stock 127.41903
#R| 5208 2020 391 550 quality 85.68508
#R| 5209 2020 284 229 stock 95.69629
#R| 5210 2020 440 853 quality 92.31113Wr Summary Data Frame
Summary statistics of relative weight for all fish is computed below.
wr_all <- dat |>
group_by(year) |>
summarize(n=n(),
mn_wr=mean(wr,na.rm=TRUE),
sd_wr=sd(wr,na.rm=TRUE),
se_wr=FSA::se(wr,na.rm=TRUE))
FSA::headtail(wr_all)#R| year n mn_wr sd_wr se_wr
#R| 1 2004 300 116.50388 16.405045 0.9650026
#R| 2 2005 305 103.70058 14.860984 0.8509375
#R| 3 2006 269 107.66011 20.258635 1.2351908
#R| 15 2018 76 107.05779 25.899713 2.9709008
#R| 16 2019 219 97.20635 7.485436 0.5058189
#R| 17 2020 198 94.78333 8.648857 0.6146475Summary statistics of relative weight for each length category is computed below.
wr_gcat <- dat |>
group_by(year,gcat) |>
summarize(n=n(),
mn_wr=mean(wr,na.rm=TRUE),
sd_wr=sd(wr,na.rm=TRUE),
se_wr=FSA::se(wr,na.rm=TRUE)) |>
ungroup()
FSA::headtail(wr_gcat)#R| year gcat n mn_wr sd_wr se_wr
#R| 1 2004 substock 110 103.54857 13.477955 1.3545854
#R| 2 2004 stock 130 123.09751 11.535665 1.0117447
#R| 3 2004 quality 34 127.50055 12.158543 2.0851728
#R| 68 2020 stock 52 95.40331 10.326851 1.4320766
#R| 69 2020 quality 132 94.78427 8.119609 0.7067213
#R| 70 2020 preferred 13 92.28008 6.826683 1.8933812McCarrick et al. (2022) only plotted fish in the “stock”, “quality”, and “preferred” length categories in Figure 4; thus, only these length categories are retained below.
wr_gcat <- wr_gcat |>
filter(gcat %in% c("stock","quality","preferred"))The summaries for all fish and fish by length categories are row-bound (i.e., stacked) together to form an overall summary data frame. Because the data frame for all fish did not have a gcat variable, that variable will be populated with NA when the two data frames are bound. The ifelse() below converts these NA values to All, before gcat is factored with levels ordered as they would appear in Figure 4 and with more descriptive labels. Finally, for aesthetic purposes, I moved gcat to be the first variable and sorted the results by year within length category.
wr_dat <- bind_rows(wr_all,wr_gcat) |>
mutate(gcat=ifelse(is.na(gcat),"All",as.character(gcat)),
gcat=factor(gcat,levels=c("All","stock","quality","preferred"),
labels=c("All fish","Stock to Quality",
"Quality to Preferred","Preferred to Memorable"))) |>
relocate(gcat) |>
arrange(gcat,year)
FSA::headtail(wr_dat)#R| gcat year n mn_wr sd_wr se_wr
#R| 1 All fish 2004 300 116.50388 16.405045 0.9650026
#R| 2 All fish 2005 305 103.70058 14.860984 0.8509375
#R| 3 All fish 2006 269 107.66011 20.258635 1.2351908
#R| 66 Preferred to Memorable 2018 22 98.79180 17.476556 3.7260144
#R| 67 Preferred to Memorable 2019 17 98.61326 8.666677 2.1019780
#R| 68 Preferred to Memorable 2020 13 92.28008 6.826683 1.8933812Finally, the lower and upper confidence intervals for each mean are added to the data frame using normal distribution theory.4
4 qt() returns a the 97.5th percent critical value from a t-distribution with df degrees-of-freedom.
wr_dat <- wr_dat |>
mutate(lci=mn_wr-qt(0.975,df=n-1)*se_wr,
uci=mn_wr+qt(0.975,df=n-1)*se_wr)
FSA::headtail(wr_dat)#R| gcat year n mn_wr sd_wr se_wr lci
#R| 1 All fish 2004 300 116.50388 16.405045 0.9650026 114.60482
#R| 2 All fish 2005 305 103.70058 14.860984 0.8509375 102.02610
#R| 3 All fish 2006 269 107.66011 20.258635 1.2351908 105.22820
#R| 66 Preferred to Memorable 2018 22 98.79180 17.476556 3.7260144 91.04313
#R| 67 Preferred to Memorable 2019 17 98.61326 8.666677 2.1019780 94.15727
#R| 68 Preferred to Memorable 2020 13 92.28008 6.826683 1.8933812 88.15476
#R| uci
#R| 1 118.4029
#R| 2 105.3750
#R| 3 110.0920
#R| 66 106.5405
#R| 67 103.0693
#R| 68 96.4054This data frame, now called wr_dat, is ready for recreating Figure 4.
Recreating Figure 4
Figure 4 is a simple bar plot facetted across years similar to the PSD plot in this previous post. Confidence intervals are added as described in that same post. Thus, many of the details below are not discussed further here. However, there are two new things to consider here.
First, the y-axis in Figure 4 in (mccarricketal_2020?) is limited from 70 to 140. If limits=c(70,140) is used in scale_y_continuous() as described in this post the bars will be removed because their “data” extends to zero.5 Thus, the limits of the y-axis must be set with ylim= in coord_cartesian() to preserved the bars.
5 Data points outside axis limits created in this way are treated as missing and are removed from the figure.
Second, the y-axis label in Figure 4 in (mccaricketal_2020?) is italicized with a subscript “r”. In the markdown language, asterisks are used to denoted italics and the tilde is used to create a subscript, which can be seen in name= in scale_y_continuous(). However, to make this markdown code render properly axis.title.y= in theme must be set to element_markdown() from ggtext.
ggplot(data=wr_dat,mapping=aes(x=year,y=mn_wr)) +
geom_errorbar(mapping=aes(ymin=lci,ymax=uci),width=0.25) +
geom_col(color="black",fill="gray70",width=1) +
geom_text(mapping=aes(label=gcat),x=Inf,y=Inf,vjust=1.25,hjust=1.05,size=3,
check_overlap=TRUE) +
scale_y_continuous(name="*W~r~*",expand=expansion(mult=0),
breaks=scales::breaks_width(10)) +
scale_x_continuous(name="Year",
limits=c(2000,2022),breaks=scales::breaks_width(2),
expand=expansion(mult=0)) +
coord_cartesian(ylim=c(70,140)) +
lemon::facet_rep_wrap(vars(gcat),ncol=1) +
theme_bw() +
theme(panel.grid=element_blank(),
strip.background=element_blank(),
strip.text=element_blank(),
axis.title.y=ggtext::element_markdown())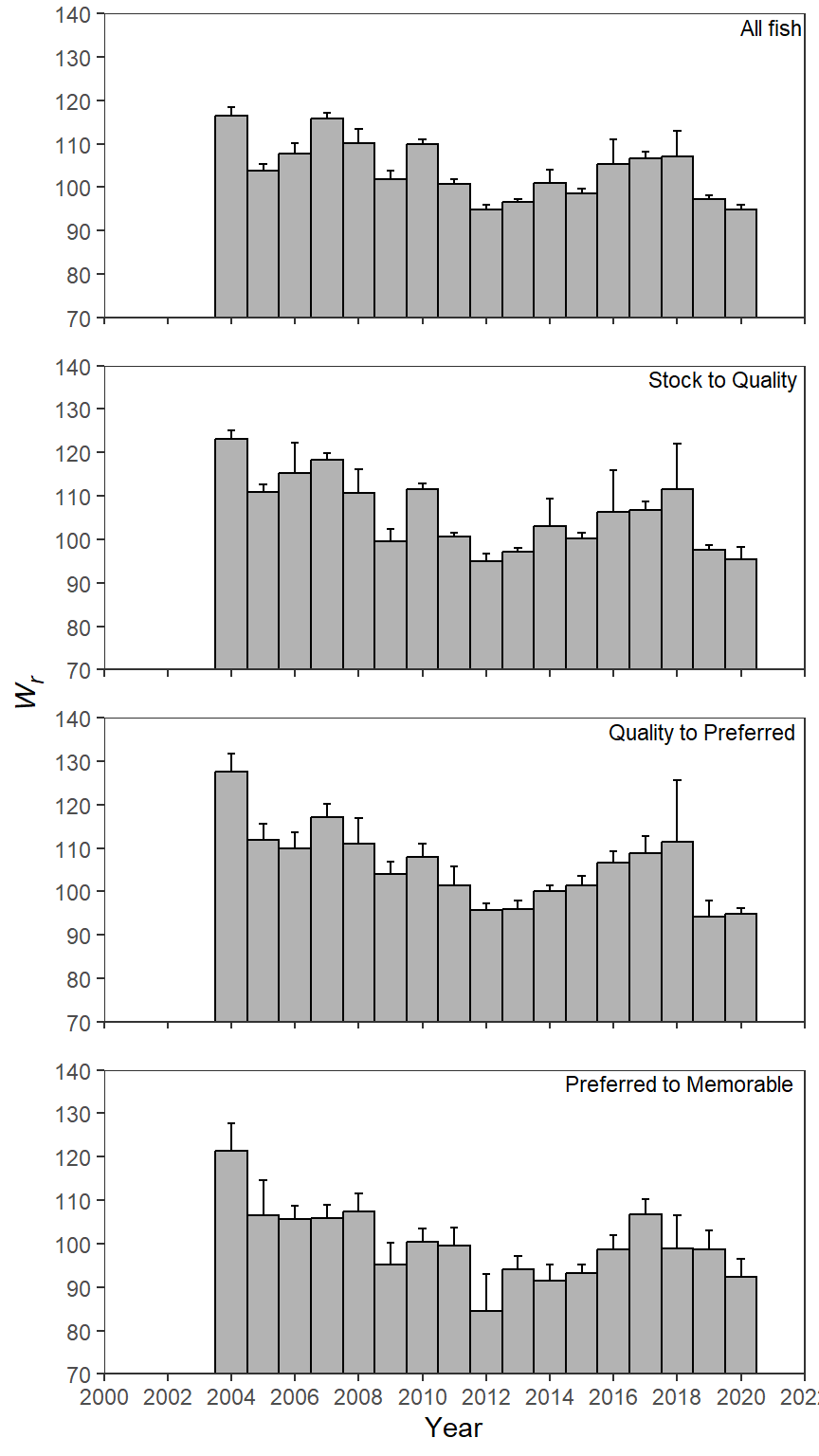
Further Thoughts
Point-and-Lines Plot
As mentioned in this previous post I understand that these are the much derided “dynamite plots”. Personally, I find the bars distracting (so much gray with little purpose and starting at 70) and find a point-and-lines plot, possibly with a horizontal line at the common reference value of 100.
ggplot(data=wr_dat,mapping=aes(x=year,y=mn_wr)) +
geom_errorbar(mapping=aes(ymin=lci,ymax=uci),width=0.25) +
geom_line(linewidth=0.75,color="gray70") +
geom_point(size=1) +
geom_hline(yintercept=100,color="gray30",linetype="dashed",linewidth=0.5) +
geom_text(mapping=aes(label=gcat),x=Inf,y=Inf,vjust=1.25,hjust=1.05,size=3,
check_overlap=TRUE) +
scale_y_continuous(name="*W~r~*",limits=c(70,140),expand=expansion(mult=0),
breaks=scales::breaks_width(10)) +
scale_x_continuous(name="Year",
limits=c(2000,2022),breaks=scales::breaks_width(2),
expand=expansion(mult=0)) +
lemon::facet_rep_wrap(vars(gcat),ncol=1) +
theme_bw() +
theme(panel.grid=element_blank(),
strip.background=element_blank(),
strip.text=element_blank(),
axis.title.y=ggtext::element_markdown())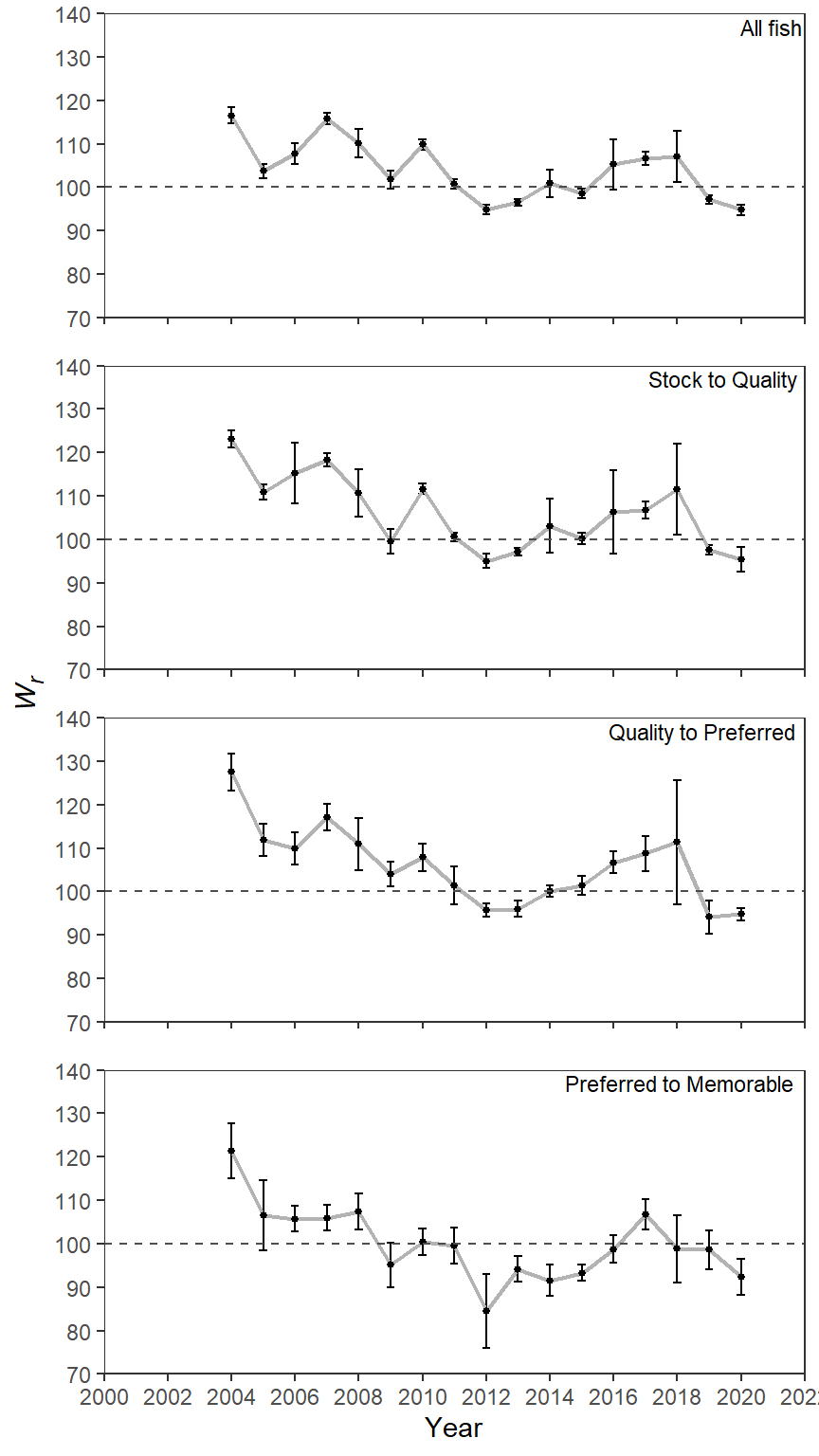
References
Reuse
Citation
@online{h._ogle2023,
author = {H. Ogle, Derek},
title = {McCarrick Et Al. (2022) {Wr} {Plot}},
date = {2023-03-26},
url = {https://fishr-core-team.github.io/fishR/blog/posts/2023-3-26_McCarricketal2022_Fig4/},
langid = {en}
}